Diagnosis
Computer-assisted medical diagnosis has long been a goal of the Artificial Intelligence community. In the 1960’s and 1970’s, the enabling technology for this was to have been Bayesian probability analysis. However, although human diseases could be represented (or modeled) efficiently as Bayesian networks, it was soon discovered that these models were too big to analyze – either exactly or by approximation. Intensive research over the last fifty years has produced improvements in analytical methods for these networks, but has failed to produce a system capable of diagnosing human disease.
This has led to a number of alternative, non-Bayesian approaches to dealing with probabilities including: various statistical weighting schemes, expert systems, and fuzzy logic. All these approaches have had to rely on highly simplified models of diseases and approximations that were inaccurate and unreliable. The combination of these factors has made existing diagnostic products so inaccurate that they have experienced little acceptance in clinical use (for example, consider the current spate of “symptom checkers”).
Over the last twenty-five years, we have researched this problem and developed a unique technology for analyzing large Bayesian networks, which we believe is the most powerful in the world. This power allows us to model diseases with sufficient complexity to ensure accuracy. Although our current disease model is fairly small (a portion of infectious diseases), the technology is designed to scale up efficiently. Tests of this model and scaled-up versions of it have demonstrated both computational speed and diagnostic accuracy.
After producing a disease differential, our system then assists the deductive process of narrowing that differential by performing a sensitivity analysis to assign a value for each, as yet unknown, finding (symptom, sign, or test result). The value for each potential finding is a measure of how much the probabilities of the diseases in the differential will change once the finding is known. This value can be used to help decide what information to gather next.
To further improve the efficiency of this process, each potential finding is assigned to a diagnostic “level”. The levels correspond to increasing cost, time, risk, and skill required. By starting at the lowest level and progressing through higher levels, the clinician receives efficient, cost-effective guidance for the diagnostic process. Significantly, expensive diagnostic tests that add little to the diagnostic process can be avoided. This level-based approach to diagnosis also allows more of the work to be performed by health care workers with less training than physicians, a benefit that can help address the existing and worsening shortage of primary care physicians worldwide.
Covid-19 Example
The Bayesian network models of human disease mentioned in the previous section were developed for the CDC and were focused on quarantinable diseases and those non-quarantinable diseases that would be included in a differential diagnosis of a quarantinable disease. Since the current pandemic is caused by a quarantinable disease – Covid-19 – we have added it to our disease models.
A hypothetical case of Covid-19 will be described and diagnosed using our medical diagnostic system both when it does not contain the Covid-19 model and when it does.
Screenshots will be presented to demonstrate items being discussed. These screenshots were taken from a development and demonstration prototype. They do not represent an actual product nor the best formats for how a real product might appear. On a phone, they are best viewed horizontally.
Presentation
A fifty-year old woman presented at her clinic asserting that she had had several symptoms intermittently over the previous six days:
- Abdominal pain
- Chills
- Shortness of breath at rest (Dyspnea)
- Mild fatigue
- Headache
- Clear nasal Discharge
- Smell change
- Diarrhea
While she had felt none of these was serious enough for her to seek medical help, she now had what she felt were more serious symptoms:
- Anorexia
- Cough
- High temperature (102.6 degrees Fahrenheit)
Upon examination by a physician, additional symptoms were detected:
- Abnormal respiratory sounds were present (rales)
- Respiratory rate was increased (tachypnea)
- Lowered blood oxygen was moderate (hypoxemia)
Several laboratory tests including a Complete Blood Count (CBC) and a Comprehensive Metabolic Panel were performed and produced the following abnormalities:
- Erythrocite Sedimentation Rate was increased (ESR)
- Platelets were low (122)
- White blood cell count was low (3.6)
- Lymphocyte count was decreased (0)
- AST was elevated
- Bilirubin total was elevated
First, the patient will be diagnosed as if she was one of the first few cases of covid-19 when doctors had no idea what they were dealing with.
Disease Differential
When these findings for day seven were evaluated using the system without the Covid-19 model, the following disease differential was produced (in part):
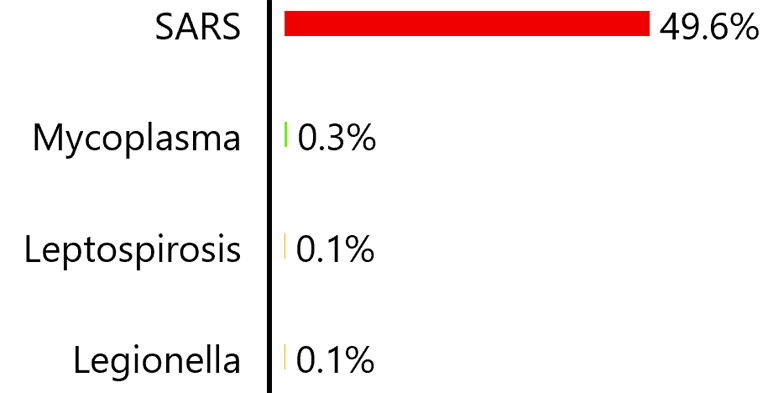
The colors on the bars indicate the severity of the disease. Red is the highest indicating a quarantinable disease, then shades down through orange and yellow to green indicating a mild disease like the common cold.
Next, the patient will be diagnosed as if it was December 1, 2020, when Covid-19 was fairly well understood. This is the date of the disease model used with this example.
Disease Differential
When these findings for day seven were evaluated using the system with the Covid-19 model, the following disease differential was produced (in part):
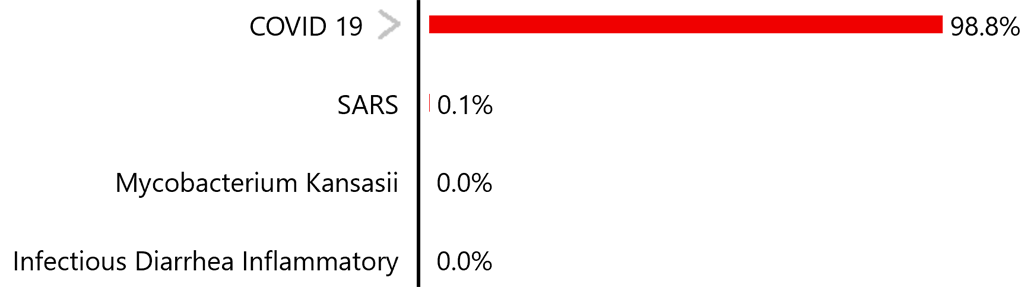
The greater-than symbol indicates the disease has different types which can be viewed by tapping the symbol:
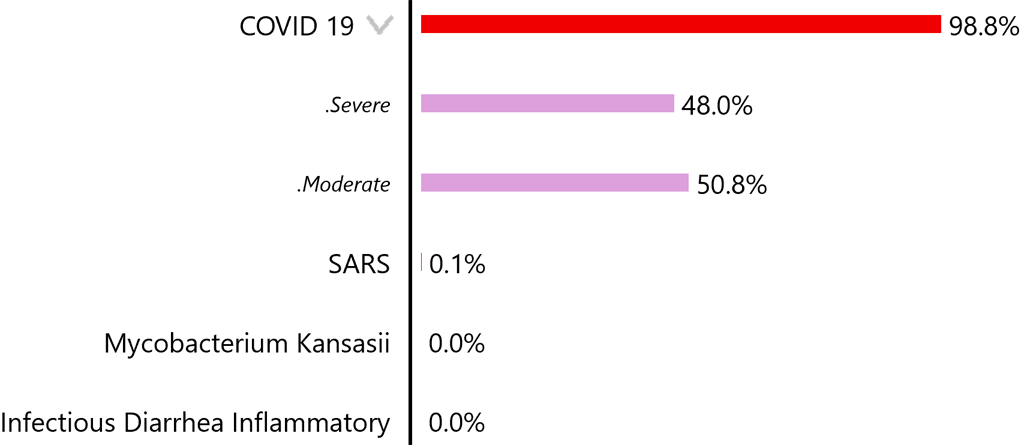
With the current pandemic focus on Covid-19, it is unlikely that many cases of it will not be diagnosed. There is, however, a serious possibility of other diseases sharing symptoms with Covid-19, being diagnosed incorrectly as Covid-19. Allowing a disease like SARS to “fall through the crack” is something that could be prevented with the use of an unbiased diagnostic tool where every disease competes equally to account for a presentation.
Emerging Disease
When these findings for day seven of the disease were evaluated using the system without the Covid-19 model, it produced the “weak” disease differential shown above – the highest disease likelihood was only 49.6%. As the disease progressed though succeeding days, it would fail to produce a consistently strong diagnosis of a single disease. For each day, the diagnostic process would try to determine the answer to the following question, “Which disease is most likely to have produced the symptoms and test results experienced by the patient?” Since no known disease’s progression would match that of Covid-19, the answer to the question would differ from day to day.
Ultimately, the sequence will have answered a different question, “Is the presentation simply an outlier for a known disease in the disease models and so is difficult to diagnose for that reason, or is the disease not in the disease models?” This question is answered by the nature of subsequent cases. Since an “outlier” is by definition a rare event, it is highly unlikely to occur again. However, if a series of undiagnosable presentations occur within a fairly short time, it is highly likely to be an emerging disease. In that case, public health actions can be initiated quickly to contain the disease and possibly avoid a new pandemic.
Continuing the case presented above, we will follow a 30-day progression of the disease, once with the disease model containing Covid-19 and once without it. The simulation will employ the same “doctor” for each progression. The “doctor” will obtain a differential for the then current set of findings, request a sensitivity analysis*, choose the unknown finding with the highest sensitivity value, employ Monte Carlo sampling to obtain a state for the finding and continue the process until no additional findings have a high-enough sensitivity value to continue. Note that random sampling is consistent between the two progressions and that no interventions are attempted.
* sensitivity analysis – A process wherein a heuristic value is calculated for each unknown finding. The value is a measure of how much knowing the state of a finding affects the probabilities of the diseases in the differential. The higher the changes in probabilities, the greater the value.
As the disease progresses, the findings it is likely to affect will change. For example, some findings will be more likely to occur during the early stages while others will be more likely to occur during later stages. When the Covid-19 model is present, the model tracks these changes well and the diagnosis remains fairly stable:
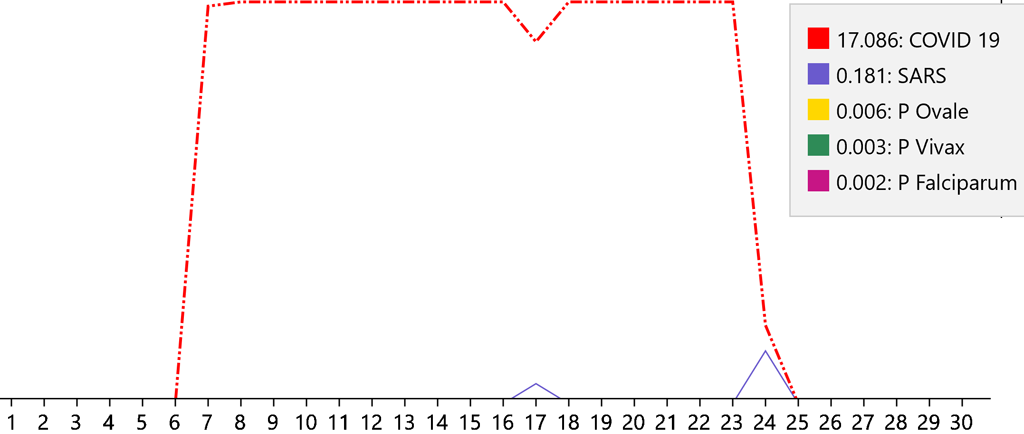

The numbers on the base line are the days of the disease. The points on each line occur at the probabilities of a disease on each day. So, it can be seen that the patient presented on day seven when the diagnosis was initiated. Also, it can be seen that the disease completed its course on day twenty-five. The ranking of the five highest diseases are graphed. The legend shows the rank of the diseases together with the color used to graph it. The highest-ranking disease is graphed with a dashed red line. The ranking number is the area of the graph under the line for that disease. Day seventeen shows the change due to a change in the value of Fibrinogen.
When the Covid-19 model is not present, the day-to-day changes in the disease cause different diseases to be diagnosed:
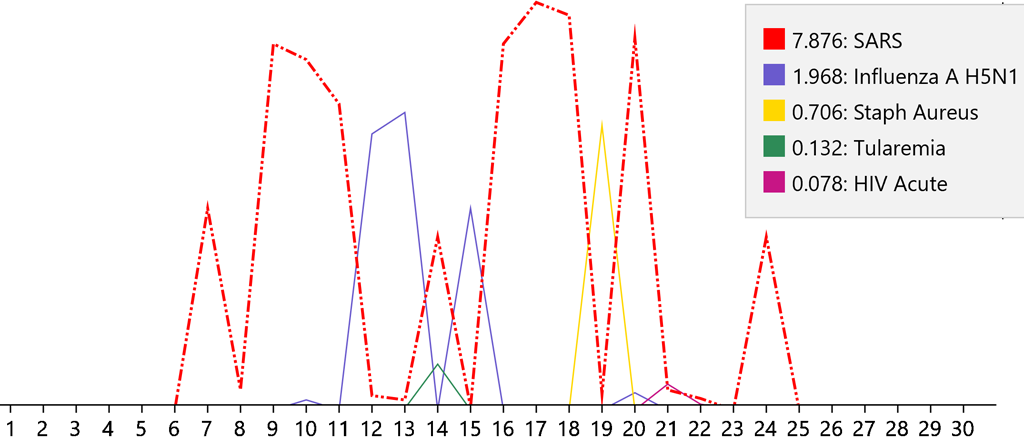

This erratic diagnostic pattern shows that the changes that occur over time do not match those of a known disease. Consequently, it must be unknown and therefore an emerging disease. Note that the diagnosis of SARS most closely follows that of Covid-19 due to the similarity of the viruses – SARS-CoV and SARS-CoV-2.
Although the inconsistency of diagnoses without Covid-19 in the disease models would indicate an emerging disease, there would still be some consistency in the presentations; e.g., fevers, shortness of breath, and other Covid-19 symptoms. Searching for other undiagnosable cases sharing those symptoms in other locations could indicate that the disease had already spread beyond the location where it was first detected.
Imagine how different the world would be today if enough undiagnosable cases of Covid-19 had been detected in Wuhan, China, to identify it as an emerging disease when it first appeared – and the current pandemic had been avoided!
Philanthopic Aquisition
For a system that could detect emerging diseases to be maximally effective, it should be in global use and by the most people possible. That would suggest that the system should be provided by an independent organization (for example the WHO) and offered free of charge in order to be accessible to those in the developing world many of whom cannot afford to pay for healthcare, but who might unknowingly be helping to promote the development of new diseases; for example by buying and eating “wild” meat. This should also suggest that any profit-oriented ownership must be avoided.
In consequence, we have decided to offer our technology for a philanthropic acquisition:
- The technology would be placed in the public domain without patenting or other restrictions and so could be used freely by anyone.
- An additional endowment would support a non-profit organization such as the one mentioned in the preceding paragraph, that would develop, maintain and deploy the system.
The acquisition would not only provide a timely defense against new pandemics, but would also provide the world population with efficient, accurate medical diagnosis. This would have a significant impact on global health with accompanying social and economic benefits. With the projected global cost of the current pandemic in the trillions, such a system would certainly be worth billions. At a billion, it would be a bargain.
If you are interested in making the acquisition or significantly contributing to it, please send an email to Acquisition@ArchipelagoSystems.net. Note that we will not respond to venture capital or other forms of private ownership.